- Joined
- Nov 5, 2016
- Messages
- 148
- Reaction score
- 30
Grabbed another scientific journal article from the AAMC Section Bank. This one was from C/P passage 9. The red text in this post are my questions that maybe one of yall can answer/clarify for me! Feel free to devour this article yourself and compare your thoughts to mine to see if we differ at all. (I'm only reading the parts of the journal article that pertain to the AAMC SB C/P passage 9 in an attempt to make this study strategy more time efficient, so I'd recommend bringing that up to make sure you're not spending too much time on this article).
MCAT material: AAMC Section Bank C/P passage 9
Experimental-based passage skills that AAMC apparently wants us to know:
1. Analyzing folded molecules (ie. folded RNA) via Electrophoresis
2. Analyzing a Thermal Denaturation (Tm) graph
Article source: Secondary Structure of a Conserved Domain in an Intron of Influenza A M1 mRNA
Title: Secondary Structure of a Conserved Domain in an Intron of Influenza A M1 mRNA
(Important background info for this article:
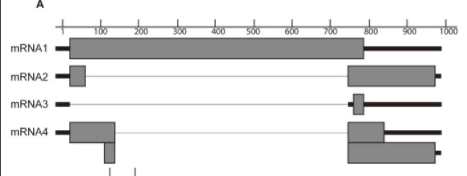
^This is showing Influenza A virus's pre-mRNA segment 7 and how it can be alternatively spliced into M1, M2, M3, and M4 mRNAs)
What’s the big picture
Influenza A virus is quite a dangerous - and sometimes fatal - virus. Currently, we don’t have many drugs to treat this virus. We must develop new antiviral treatments as well as more effective vaccines.
Main idea/gist of the article
Influenza A virus’s RNA structure plays a key role for this virus (ie. determining protein expression of its various, necessary proteins). In particular, Segment 7 of the vRNA must be alternatively spliced correctly (in order to express the right proteins at the right times) over the course of infection in order for the virus to succeed in infecting its host.
Purpose of the study
To better understand the structural characteristics of the Influenza A viral (RNA) genome (so we can make better treatments/vaccines for this virus).
The scientific logic/strategy behind each trial or experiment (e.g., Why did they choose those conditions? What variables were they controlling or isolating? What did they hope to clarify?)
(General info about the previously hypothesized structure of a particular region of M1 mRNA (maybe rotate your head sideways to see it better lol):
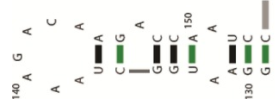
^M1 mRNA hairpin structure was hypothesized previously (again, only a section of the M1 mRNA is shown here).)
Experiment: Native Gel Electrophoresis and Enzymatic/Chemical Mapping
Study purpose - to determine if the M1 mRNA region in question forms a a) multi-branch structure -or- b) a hairpin structure (at nucleotides 127-170, which is an important location on this mRNA since we previously mentioned that this region of the mRNA is highly conserved/stable for Influenza A viruses).
IV’s:
RNA
Wild Type RNA
Mutant RNA designed to form the multi-branch structure (MBmutant)
Mutant RNA designed to form the hairpin structure (HPmutant)
Denatured WT RNA, MBmutant RNA, and HPmutant RNA (control variable)
DV:
how far each RNA travels on a gel (indicates which structure the WT resembles).
Results (Figure A was the native RNA conformations & Figure B was the denatured RNA conformations):

(^Translation of Figure A into words: “The Wild Type RNA resembles the MBmutant, not the HPmutant”)

minus
(^Translation of Figure B into words: “all of the RNA’s used were of the same length”; this is essentially a control gel)
^Conclusion:
The Wild Type RNA forms the b) multi-branch structure.
The M1 mRNA - at nucleotides 127-190 - forms a multi-branch structure, not a hairpin structure.
Experiment: Optical Melting Experiments Reveal Similar Melting Profiles in 1 M NaCl and in 10 mM MgCl2
Study purpose - to determine what type (and what concentration) of salt (ie. KCl or NaCl) to include in our Mg2+ experimental buffer when experimenting with the RNA secondary structures?
IV’s:
Wild-Type RNA
Temperature (increasing)
Tx of Wild-Type RNA
100 mM KCl without MgCl2 (the blue line in the graph)
100 mM KCl with 10mM MgCl2 (the black line in the graph)
300 mM KCl with 10mM MgCl2 (the green line in the graph)
1 M NaCl (without MgCl2) (the red line in the graph)
DV:
absorbance at 280 nm (absorbance indicates that this reaction has occurred: native RNA → denatured RNA... explanation of exactly how this is?)
Results:

^Conclusion:
-KCl alone resulted in RNA denaturing at a lower Temperature (58 degreesC).
- all other salt combos added to the RNA solution resulted in the RNA denaturing at a higher Temperature (68 degreesC).
- KCl alone denatures RNA at the lowest Temperature.
- added Mg2+ stabilizes the melting Temperature (to 68 degreesC), regardless of how much KCl is added.
- 1 M NaCl resulted in RNA denaturing at 68 degrees C.
- 1 M NaCl should be used in our buffer to predict RNA secondary structures in the presence of Mg2+.
^Can someone explain to me why we are even doing all of this and why we chose K+, Na+, and Mg2+...I thought all of these are present in our cells anyways, so why wouldn’t we include them all in our experimental buffers?
What can be concluded from the results/data?
Influenza A virus's M1 mRNA - at nucleotides 127-170 - form a multi-branch structure (rather than a hairpin structure).
Final, overall, collective thoughts
The M1 protein for Influenza A viruses is a critical protein for these viruses. By knowing more about M1 mRNA’s secondary structure, we could potentially develop better treatments/vaccines for Influenza A viral infections. This study shows that at an important region of the M1 mRNA (at nucleotides 79-190, which is highly conserved/stable among Influenza A viruses), it forms a multi-branch secondary structure rather than a hairpin structure. Knowing this, we are one step closer to potentially developing a treatment and/or vaccine for Influenza viral infections.
MCAT material: AAMC Section Bank C/P passage 9
Experimental-based passage skills that AAMC apparently wants us to know:
1. Analyzing folded molecules (ie. folded RNA) via Electrophoresis
2. Analyzing a Thermal Denaturation (Tm) graph
Article source: Secondary Structure of a Conserved Domain in an Intron of Influenza A M1 mRNA
Title: Secondary Structure of a Conserved Domain in an Intron of Influenza A M1 mRNA
(Important background info for this article:
^This is showing Influenza A virus's pre-mRNA segment 7 and how it can be alternatively spliced into M1, M2, M3, and M4 mRNAs)
What’s the big picture
Influenza A virus is quite a dangerous - and sometimes fatal - virus. Currently, we don’t have many drugs to treat this virus. We must develop new antiviral treatments as well as more effective vaccines.
Main idea/gist of the article
Influenza A virus’s RNA structure plays a key role for this virus (ie. determining protein expression of its various, necessary proteins). In particular, Segment 7 of the vRNA must be alternatively spliced correctly (in order to express the right proteins at the right times) over the course of infection in order for the virus to succeed in infecting its host.
Purpose of the study
To better understand the structural characteristics of the Influenza A viral (RNA) genome (so we can make better treatments/vaccines for this virus).
The scientific logic/strategy behind each trial or experiment (e.g., Why did they choose those conditions? What variables were they controlling or isolating? What did they hope to clarify?)
(General info about the previously hypothesized structure of a particular region of M1 mRNA (maybe rotate your head sideways to see it better lol):

^M1 mRNA hairpin structure was hypothesized previously (again, only a section of the M1 mRNA is shown here).)
Experiment: Native Gel Electrophoresis and Enzymatic/Chemical Mapping
Study purpose - to determine if the M1 mRNA region in question forms a a) multi-branch structure -or- b) a hairpin structure (at nucleotides 127-170, which is an important location on this mRNA since we previously mentioned that this region of the mRNA is highly conserved/stable for Influenza A viruses).
IV’s:
RNA
Wild Type RNA
Mutant RNA designed to form the multi-branch structure (MBmutant)
Mutant RNA designed to form the hairpin structure (HPmutant)
Denatured WT RNA, MBmutant RNA, and HPmutant RNA (control variable)
DV:
how far each RNA travels on a gel (indicates which structure the WT resembles).
Results (Figure A was the native RNA conformations & Figure B was the denatured RNA conformations):

(^Translation of Figure A into words: “The Wild Type RNA resembles the MBmutant, not the HPmutant”)

minus
(^Translation of Figure B into words: “all of the RNA’s used were of the same length”; this is essentially a control gel)
^Conclusion:
The Wild Type RNA forms the b) multi-branch structure.
The M1 mRNA - at nucleotides 127-190 - forms a multi-branch structure, not a hairpin structure.
Experiment: Optical Melting Experiments Reveal Similar Melting Profiles in 1 M NaCl and in 10 mM MgCl2
Study purpose - to determine what type (and what concentration) of salt (ie. KCl or NaCl) to include in our Mg2+ experimental buffer when experimenting with the RNA secondary structures?
IV’s:
Wild-Type RNA
Temperature (increasing)
Tx of Wild-Type RNA
100 mM KCl without MgCl2 (the blue line in the graph)
100 mM KCl with 10mM MgCl2 (the black line in the graph)
300 mM KCl with 10mM MgCl2 (the green line in the graph)
1 M NaCl (without MgCl2) (the red line in the graph)
DV:
absorbance at 280 nm (absorbance indicates that this reaction has occurred: native RNA → denatured RNA... explanation of exactly how this is?)
Results:

^Conclusion:
-KCl alone resulted in RNA denaturing at a lower Temperature (58 degreesC).
- all other salt combos added to the RNA solution resulted in the RNA denaturing at a higher Temperature (68 degreesC).
- KCl alone denatures RNA at the lowest Temperature.
- added Mg2+ stabilizes the melting Temperature (to 68 degreesC), regardless of how much KCl is added.
- 1 M NaCl resulted in RNA denaturing at 68 degrees C.
- 1 M NaCl should be used in our buffer to predict RNA secondary structures in the presence of Mg2+.
^Can someone explain to me why we are even doing all of this and why we chose K+, Na+, and Mg2+...I thought all of these are present in our cells anyways, so why wouldn’t we include them all in our experimental buffers?
What can be concluded from the results/data?
Influenza A virus's M1 mRNA - at nucleotides 127-170 - form a multi-branch structure (rather than a hairpin structure).
Final, overall, collective thoughts
The M1 protein for Influenza A viruses is a critical protein for these viruses. By knowing more about M1 mRNA’s secondary structure, we could potentially develop better treatments/vaccines for Influenza A viral infections. This study shows that at an important region of the M1 mRNA (at nucleotides 79-190, which is highly conserved/stable among Influenza A viruses), it forms a multi-branch secondary structure rather than a hairpin structure. Knowing this, we are one step closer to potentially developing a treatment and/or vaccine for Influenza viral infections.
Last edited:
