- Joined
- Jun 20, 2016
- Messages
- 264
- Reaction score
- 49
Native-page: everything in-tact, separated by size and charge or shape.
ie. If we had a heterotrimer, we only would see one band.
SDS-page non-reducing(w/o B-ME): S-S are intact but protein is denatured
ie. If we had a heterotrimer, we would only see one band.
SDS-page reducing (w/ B-ME): S-S become reduced protein is still dentured.
ie. If we had a heterotrimer, we would see 3 separate bands.
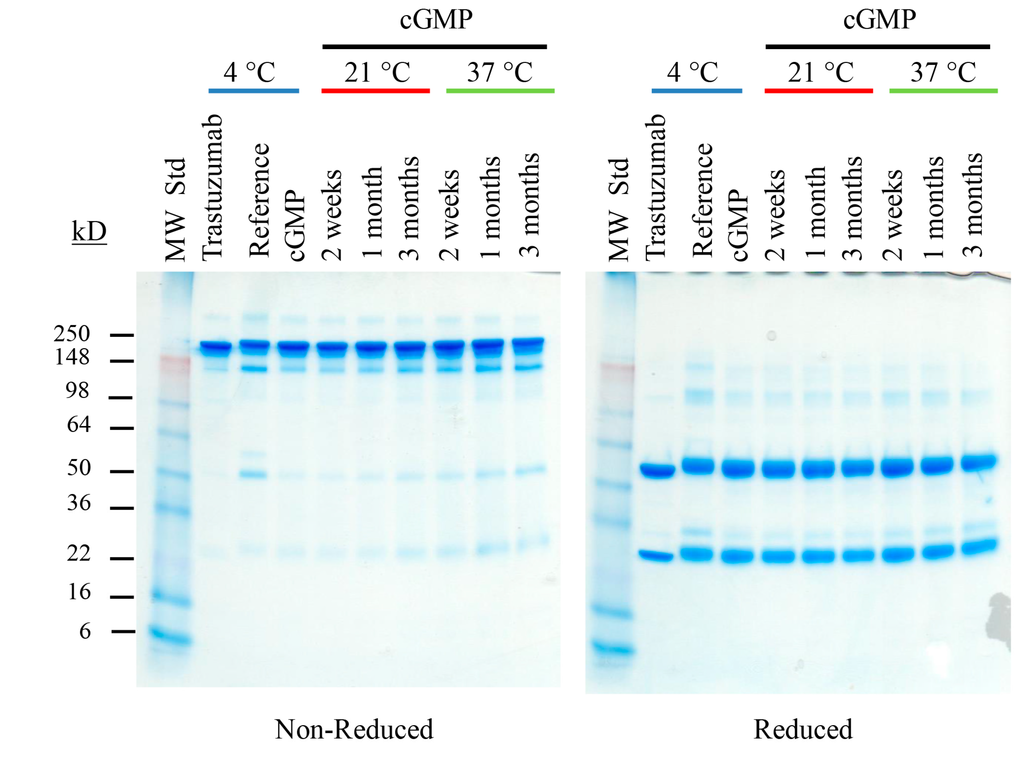
From this picture, we can say that the protein was a heterodimer (or whatever 'mer' in this case, just it can create 2 different KDa of subunits)

In this picture, I notice that the non-reducing detected higher MW proteins than reducing. Looking at lane 5, what can we deduce? If it were to be a homodimer, then I think I would only see one band at around 7kDa and not 18.4.
I guess what is the main difference between native-page and SDS-page non-reduced?
The only thing I can think of for now is that if you run a heterodimer under both.
If you see one band for native-page and 2 bands for SDS-page, then the dimers weren't connected by a disulfide bridge.
ie. If we had a heterotrimer, we only would see one band.
SDS-page non-reducing(w/o B-ME): S-S are intact but protein is denatured
ie. If we had a heterotrimer, we would only see one band.
SDS-page reducing (w/ B-ME): S-S become reduced protein is still dentured.
ie. If we had a heterotrimer, we would see 3 separate bands.

From this picture, we can say that the protein was a heterodimer (or whatever 'mer' in this case, just it can create 2 different KDa of subunits)

In this picture, I notice that the non-reducing detected higher MW proteins than reducing. Looking at lane 5, what can we deduce? If it were to be a homodimer, then I think I would only see one band at around 7kDa and not 18.4.
I guess what is the main difference between native-page and SDS-page non-reduced?
The only thing I can think of for now is that if you run a heterodimer under both.
If you see one band for native-page and 2 bands for SDS-page, then the dimers weren't connected by a disulfide bridge.
