Note: Question at bottom in blue.
In DNA replication the DNA polymerase reads the template strand 3'->5' (or synthesizes the new DNA strand 5'->3', whichever you prefer) starting after the primer. But at the very end of the lagging strand the final okazaki fragment either 1) can't be primed (due to the primer sequence not being the absolute last DNA bases of the end of the strand) or 2) the terminal primer can't be converted into DNA (b/c the typical DNA polymerases add DNA after the primer, and the DNA polymerases that replace the primer with DNA nucleotides require a 3' strand extending beyond the primer, which obviously is not present).
To remedy this, Eukaryotes have telomeres, telomerase, etc. to prevent the shortening of the DNA strands. Prokaryotes have ciruclar DNA that doesn't have this problem.
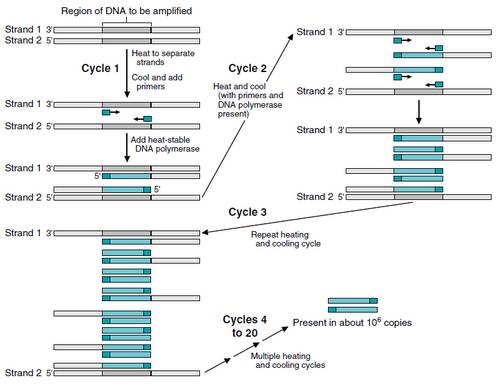
All the above to understand where I'm coming from: Why, in PCR, are the target strands of successive DNA replications not shortened? What am I missing in the PCR mechanism?
In DNA replication the DNA polymerase reads the template strand 3'->5' (or synthesizes the new DNA strand 5'->3', whichever you prefer) starting after the primer. But at the very end of the lagging strand the final okazaki fragment either 1) can't be primed (due to the primer sequence not being the absolute last DNA bases of the end of the strand) or 2) the terminal primer can't be converted into DNA (b/c the typical DNA polymerases add DNA after the primer, and the DNA polymerases that replace the primer with DNA nucleotides require a 3' strand extending beyond the primer, which obviously is not present).
To remedy this, Eukaryotes have telomeres, telomerase, etc. to prevent the shortening of the DNA strands. Prokaryotes have ciruclar DNA that doesn't have this problem.
All the above to understand where I'm coming from: Why, in PCR, are the target strands of successive DNA replications not shortened? What am I missing in the PCR mechanism?

 ) But ya you ask a valid question. The primer portion of the PCR amplification doesn't get amplified at all.
) But ya you ask a valid question. The primer portion of the PCR amplification doesn't get amplified at all.