just realized i didnt really ask a specific question lol
what are RFLPs? do we even need to know them for the genetics section?
Restriction Fragment Length Polymorphism (RFLP)
Introduction
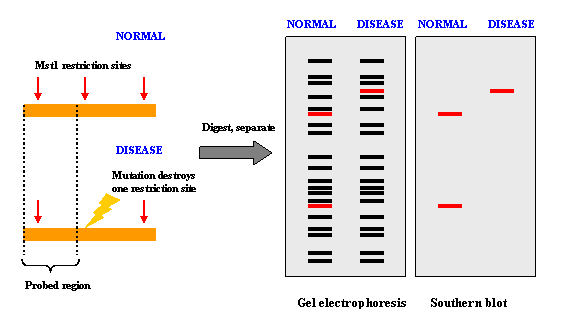
Restriction Fragment Length Polymorphism (RFLP) is a difference in homologous DNA sequences that can be detected by the presence of fragments of different lengths after digestion of the DNA samples in question with specific
restriction endonucleases. RFLP, as a molecular marker, is specific to a single clone/restriction enzyme combination.
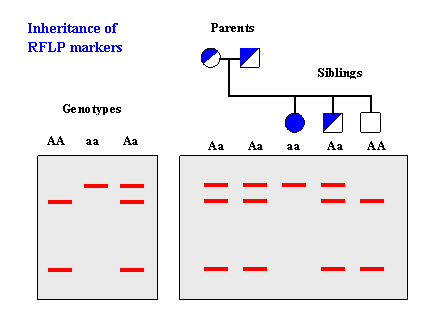
Most RFLP markers are co-dominant (both alleles in heterozygous sample will be detected) and highly locus-specific.
An RFLP probe is a labeled DNA sequence that hybridizes with one or more fragments of the digested DNA sample after they were separated by gel electrophoresis, thus revealing a unique blotting pattern characteristic to a specific genotype at a specific locus. Short, single- or low-copy genomic DNA or cDNA clones are typically used as RFLP probes.
The RFLP probes are frequently used in
genome mapping and in
variation analysis (genotyping, forensics, paternity tests, hereditary disease diagnostics, etc.).
How It Works
SNPs or
INDELs can create or abolish restriction endonuclease (RE) recognition sites, thus affecting quantities and length of DNA fragments resulting from RE digestion.
Genotyping
Developing RFLP probes
- Total DNA is digested with a methylation-sensitive enzyme (for example, PstI), thereby enriching the library for single- or low-copy expressed sequences (PstI clones are based on the suggestion that expressed genes are not methylated).
- The digested DNA is size-fractionated on a preparative agarose gel, and fragments ranging from 500 to 2000 bp are excised, eluted and cloned into a plasmid vector (for example, pUC18).
- Digests of the plasmids are screened to check for inserts.
- Southern blots of the inserts can be probed with total sheared DNA to select clones that hybridize to single- and low-copy sequences.
- The probes are screened for RFLPs using genomic DNA of different genotypes digested with restriction endonucleases. Typically, in species with moderate to high polymorphism rates, two to four restriction endonucleases are used such as EcoRI, EcoRV, and HindIII. In species with low polymorphism rates, additional restriction endonucleases can be tested to increase the chance of finding polymorphism.
PCR-RFLP
Isolation of sufficient DNA for RFLP analysis is time consuming and labor intensive. However, PCR can be used to amplify very small amounts of DNA, usually in 2-3 hours, to the levels required for RFLP analysis. Therefore, more samples can be analyzed in a shorter time. An alternative name for the technique is
Cleaved Amplified Polymorphic Sequence (CAPS) assay.
Source:
http://www.ncbi.nlm.nih.gov/projects/genome/probe/doc/TechRFLP.shtml
I hope this helps clarify some of the points that Alex made.