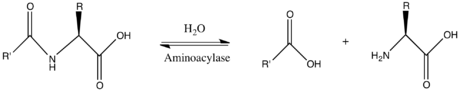
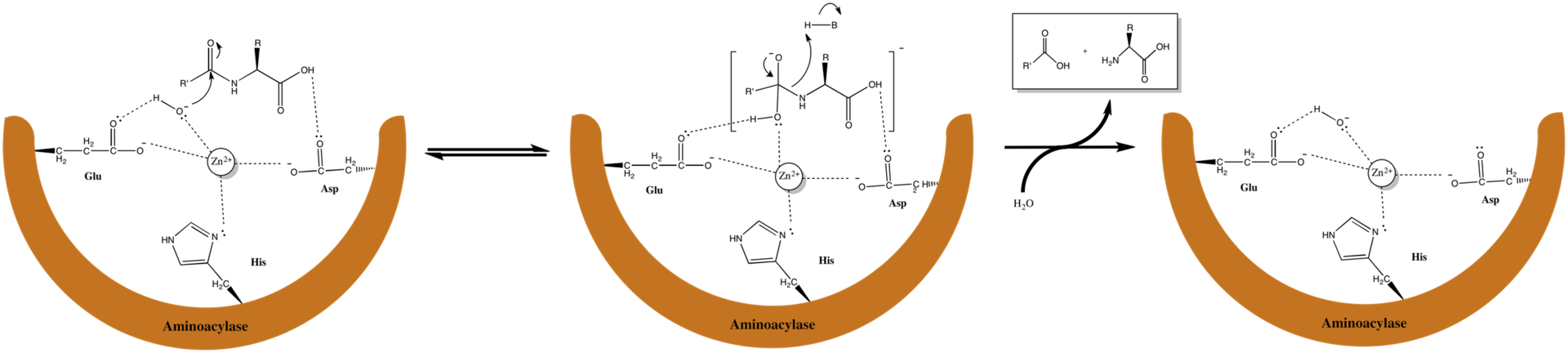
Where is the amide group in this bad boy?
Answer says water (normally in the place of glycine) is made more nucleophilic by deprotonation to make the carbonyl carbon of the amide group more electrophilic.
This might be very wrong but I think its referring to the amide and carbonyl on glutamate 148.
Edit: just kidding, glutamate definitely doesn't have an amide group
Yes it does. Because the residues are numbered, that means they are part of a polypeptide backbone, which must be linked via amide bonds. However, I'm not sure whether that's what the passage is referring to.
OP, I believe this is a zinc metalloprotease so the amide they are referring to should be the scissile amide bond, which may not be shown in this figure.